 Project Overview
Project Overview
Leak Proof CMap is a framework for the training and evaluation of phenotypic similarity methods using the Connectivity Map (CMap) L1000 transcriptomic dataset.
It establishes the most rigorous leak-proof data splitting regime reported to date, ensuring unbiased benchmarking and fair comparison of similarity methods across novel cell lines and novel mechanisms of action (MOAs).
 Technical Implementation
Technical Implementation
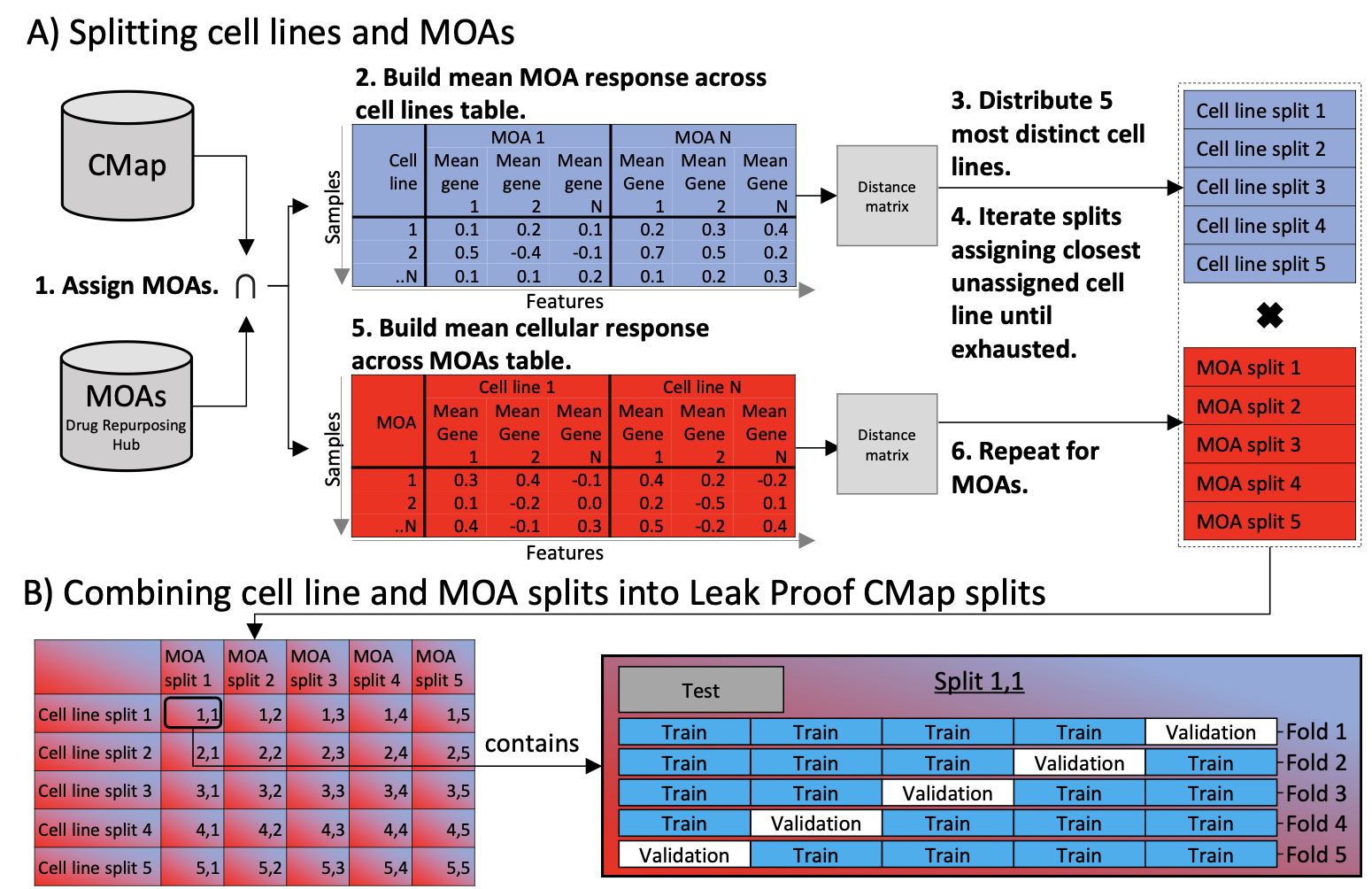
Leak Proof CMap split strategy: independent cell line and MOA splits combined into 25 leak-proof benchmark sets.
 Key Features
Key Features
-
Leak-Proof Data Splits
- Five diverse cell line splits and five MOA splits combined into 25 benchmark sets.
- Prevents information leakage between train/validation/test, mimicking deployment on unseen treatments.
-
Benchmarking Tasks
- Compactness → percent replicating metric (how tightly replicates cluster).
- Distinctness → permutation testing vs DMSO (hit calling).
- Uniqueness → AUROC (retrieval of correct replicates across large datasets).
-
Method Evaluation
- Classical metrics: Spearman Rank, Zhang, cosine, Euclidean, Euclidean-PCA.
- AI/ML method: Triplet-loss neural network embedding into 128D latent space.
- Triplet-loss significantly outperformed others in compactness and uniqueness, setting a new benchmark for similarity-based MOA discovery.
-
Open-Source Tooling
- Python package:
leakproofcmap - Includes dataset preparation, splitting routines, and benchmarking pipelines.
- Python package:
 Case Studies
Case Studies
-
Novel Cell Line Generalization
- Models evaluated on entirely unseen cell lines.
- Triplet-loss embeddings remained robust, showing potential for patient-derived samples.
-
Unbiased MOA Prediction
- Evaluation with 433 MOAs and 1,309 compounds across 30 cell lines (~177k profiles).
- Demonstrated reliable recovery of mechanisms even under strict leak-proof regimes.
-
Hit Calling in Drug Discovery
- Distinctness benchmark highlighted Euclidean-PCA as the most effective metric for hit vs control separation.
- Supports early-stage high-throughput screening campaigns.