 Project Overview
Project Overview
Phenonaut is a Python software package that revolutionizes multiomics data integration by providing a robust framework for processing and analyzing high-content imaging, proteomics, metabolomics, and other omics data. Our solution addresses critical challenges in data workflow management, including:
- Migration and version control of large datasets
- Quality control and preprocessing pipelines
- Integration of heterogeneous data types
- Auditability and reproducibility of analyses
 Technical Implementation
Technical Implementation
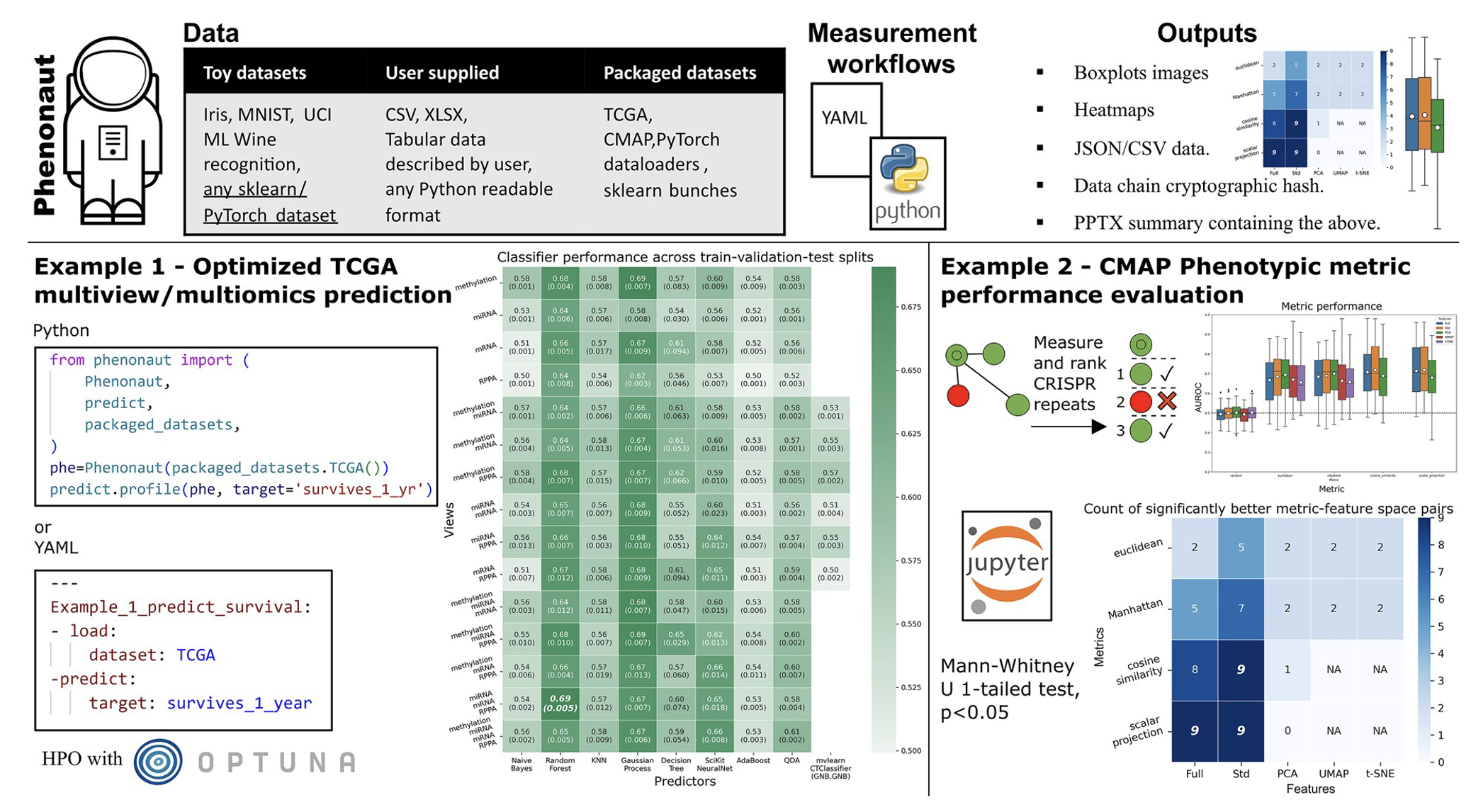 Phenonaut’s workflow visualization showing data integration and analysis pipeline
Phenonaut’s workflow visualization showing data integration and analysis pipeline
 Key Features
Key Features
-
Data Source Agnostic Integration
- Support for multiple file formats (CSV, HDF5, parquet)
- Custom data source adapters
- Automated schema validation
-
Workflow Management
- Pipeline versioning
- Checkpoint saving
- Parallel processing support
- Error handling and recovery
-
Analysis Capabilities
- Dimensionality reduction (PCA, t-SNE, UMAP)
- Statistical testing frameworks
- Visualization tools
- Custom analysis plugins
-
Quality Control
- Automated outlier detection
- Missing value handling
- Batch effect correction
- Data normalization
 Case Studies
Case Studies
-
High-Content Imaging Integration
- Processing of 100,000+ cellular images
- Feature extraction using deep learning
- Integration with proteomics data
-
Multi-omics Data Analysis
- Integration of proteomics and metabolomics
- Pathway enrichment analysis
- Network analysis
- Results: Identified novel pathway interactions